This is an old revision of the document!
Table of Contents
Objective:
To understand Gaussian process regression, and to be able to generate nonparametric regressions with confidence intervals. Also to understand the interplay between a kernel (or covariance) function, and the resulting confidence intervals of the regression.
Deliverable:
You will turn in an iPython notebook that performs Gaussian process regression on a simple dataset. You will explore multiple kernels and vary their parameter settings.
When you are done, you should produce visualizations like the following (for noiseless observations):
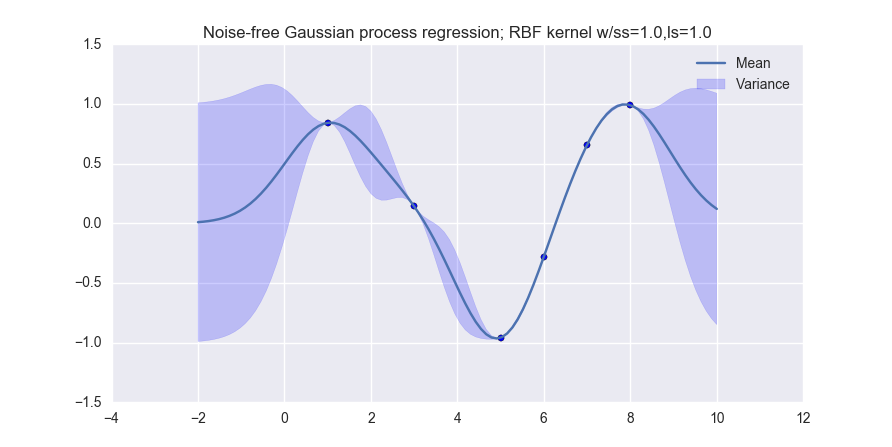
and like this (for noisy observations):
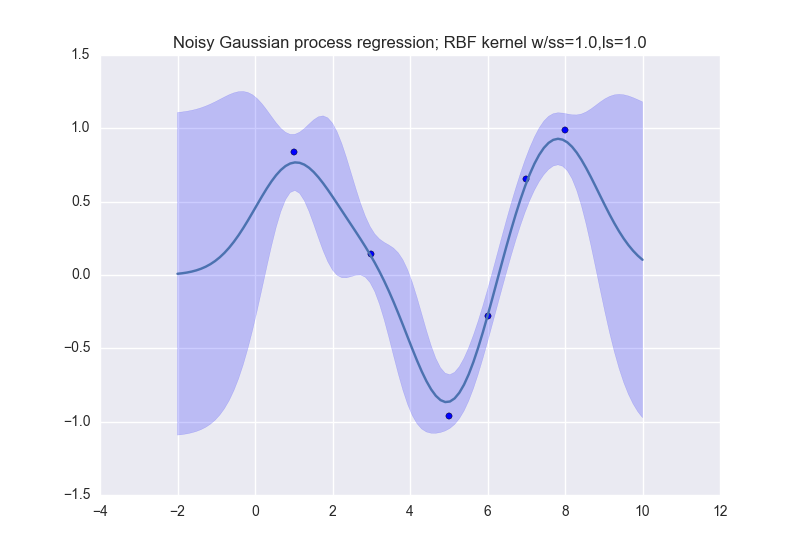
Description:
The data that we will use for this lab is simple:
data_xvals = numpy.atleast_2d( [ 1.0, 3.0, 5.0, 6.0, 7.0, 8.0 ] ) data_yvals = numpy.sin( data_xvals )
You must perform Gaussian process regression on this dataset, and produce visualizations for both noiseless and noise-free observations. Your notebook should produce one visualization for each of the following kernel types:
- The linear kernel (MLAPP 14.2.4)
- The Gaussian (or RBF) kernel (MLAPP 14.2.1; use the isotropic kernel in Eqn. 14.3)
- The polynomial kernel (MLAPP 14.2.3, in the text right after Eqn. 14.13)
Therefore, your notebook should produce six different visualizations: two for each kernel type.
For the noisy observation case, use \sigma_n^2=0.1
You should also answer the following questions:
- What happens when the bandwidth parameter \sigma_n of the Gaussian kernel gets small? Gets large?
- What happens when the degree M of the polynomial kernel gets small? Gets large?
Your visualizations should be done on the range [-2 10] of the x-axis.
Hints:
The following functions may be useful to you:
plt.gca().fill_between plt.scatter numpy.linalg.inv numpy.eye

