Table of Contents
BYU CS 501R - Deep Learning:Theory and Practice - Lab 4
Objective:
- To build a dense prediction model
- To begin reading current papers in DNN research
Deliverable:
For this lab, you will turn in a notebook that describes your efforts at
creating a pytorch radiologist. Your final deliverable is a
notebook that has (1) deep network, (2) cost
function, (3) method of calculating accuracy, (4) an image that
shows the dense prediction produced by your network on the
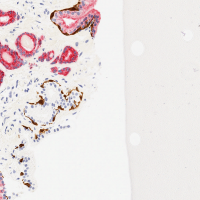
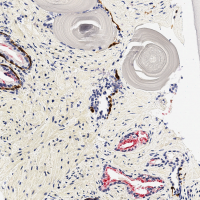
pos_test_000072.png image. This is an image in the test set that
your network will not have seen before. This image, and the ground truth labeling, is shown at the right. (And is contained in the downloadable dataset below).
Grading standards:
Your notebook will be graded on the following:
- 40% Proper design, creation and debugging of a dense prediction network
- 40% Proper implementation of a loss function and train/test set accuracy measure
- 10% Tidy visualizations of loss of your dense predictor during training
- 10% Test image output
Data set:
The data is given as a set of 1024×1024 PNG images. Each input image
(in the inputs directory) is an RGB image of a section of tissue,
and there a file with the same name (in the outputs directory) that
has a dense labeling of whether or not a section of tissue is
cancerous (white pixels mean “cancerous”, while black pixels mean “not
cancerous”).
The data has been pre-split for you into test and training splits.
Filenames also reflect whether or not the image has any cancer at all
(files starting with pos_ have some cancerous pixels, while files
starting with neg_ have no cancer anywhere). All of the data is
hand-labeled, so the dataset is not very large. That means that
overfitting is a real possibility.
The data can be downloaded here. Please note that this dataset is not publicly available, and should not be redistributed.
Description:
For a video including some tips and tricks that can help with this lab: https://youtu.be/Ms19kgK_D8w
For this lab, you will implement a virtual radiologist. You are given images of possibly cancerous tissue samples, and you must build a detector that identifies where in the tissue cancer may reside.
Part 1: Implement a dense predictor
In previous labs and lectures, we have talked about DNNs that classify an entire image as a single class. Here, however, we are interested in a more nuanced classification: given an input image, we would like to identify each pixel that is possibly cancerous. That means that instead of a single output, your network should output an “image”, where each output pixel of your network represents the probability that a pixel is cancerous.
Part 1a: Implement your network topology
Use the “Deep Convolution U-Net” from this paper: U-Net: Convolutional Networks for Biomedical Image Segmentation (See figure 1, replicated at the right). You should use existing pytorch functions (not your own Conv2D module), such as nn.Conv2d; you will also need the pytorch function torch.cat and nn.ConvTranspose2d
torch.cat allows you to concatenate tensors. nn.ConvTranspose2d is the opposite of nn.Conv2d. It is used to bring an image from low res to higher res. This blog should help you understand this function in detail.
Note that the simplest network you could implement (with all the desired properties) is just a single convolution layer with two filters and no relu! Why is that? (of course it wouldn't work very well!)
Part 1b: Implement a cost function
You should still use cross-entropy as your cost function, but you may need to think hard about how exactly to set this up – your network should output cancer/not-cancer probabilities for each pixel, which can be viewed as a two-class classification problem.
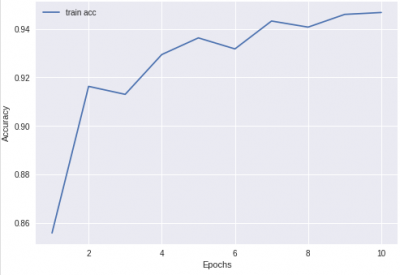
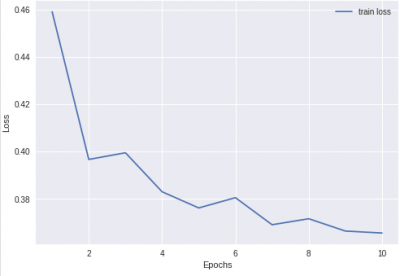
Part 2: Plot performance over time
Please generate a plot that shows loss on the training set as a function of training time. Make sure your axes are labeled!
Part 3: Generate a prediction on the pos_test_000072.png image
Calculate the output of your trained network on the pos_test_000072.png image, then make a hard decision (cancerous/not-cancerous) for each pixel. The resulting image should be black-and-white, where white pixels represent things you think are probably cancerous.
Hints:
The intention of this lab is to learn how to make deep neural nets and implement loss function. Therefore we'll help you with the implementation of Dataset. This code will download the dataset for you so that you are ready to use it and focus on network implementation, losses and accuracies.
import torchvision import os import gzip import tarfile import gc from IPython.core.ultratb import AutoFormattedTB __ITB__ = AutoFormattedTB(mode = 'Verbose',color_scheme='LightBg', tb_offset = 1) class CancerDataset(Dataset): def __init__(self, root, download=True, size=512, train=True): if download and not os.path.exists(os.path.join(root, 'cancer_data')): datasets.utils.download_url('http://liftothers.org/cancer_data.tar.gz', root, 'cancer_data.tar.gz', None) self.extract_gzip(os.path.join(root, 'cancer_data.tar.gz')) self.extract_tar(os.path.join(root, 'cancer_data.tar')) postfix = 'train' if train else 'test' root = os.path.join(root, 'cancer_data', 'cancer_data') self.dataset_folder = torchvision.datasets.ImageFolder(os.path.join(root, 'inputs_' + postfix) ,transform = transforms.Compose([transforms.Resize(size),transforms.ToTensor()])) self.label_folder = torchvision.datasets.ImageFolder(os.path.join(root, 'outputs_' + postfix) ,transform = transforms.Compose([transforms.Resize(size),transforms.ToTensor()])) @staticmethod def extract_gzip(gzip_path, remove_finished=False): print('Extracting {}'.format(gzip_path)) with open(gzip_path.replace('.gz', ''), 'wb') as out_f, gzip.GzipFile(gzip_path) as zip_f: out_f.write(zip_f.read()) if remove_finished: os.unlink(gzip_path) @staticmethod def extract_tar(tar_path): print('Untarring {}'.format(tar_path)) z = tarfile.TarFile(tar_path) z.extractall(tar_path.replace('.tar', '')) def __getitem__(self,index): img = self.dataset_folder[index] label = self.label_folder[index] return img[0],label[0][0] def __len__(self): return len(self.dataset_folder)
You are welcome to resize your input images, although don't make them so small that the essential details are blurred! I resized my images down to 512×512.
You will need to add some lines of code for memory management:
def scope(): try: #your code for calling dataset and dataloader gc.collect() print(torch.cuda.memory_allocated(0) / 1e9) #for epochs: # Call your model,loss and accuracy except: __ITB__() scope()
Since you will be using the output of one network in two places(convolution and maxpooling), you can't use nn.Sequential. Instead you will write up the network like normal variable assignment as the example shown below:
class CancerDetection(nn.Module): def __init__(self): super(CancerDetection, self).__init__() self.conv1 = nn.Conv2d(3,64,kernel_size = 3, stride = 1, padding = 1) self.relu2 = nn.ReLU() self.conv3 = nn.Conv2d(64,128,kernel_size = 3, stride = 1, padding = 1) self.relu4 = nn.ReLU() def forward(self, input): conv1_out = self.conv1(input) relu2_out = self.relu2(conv1_out) conv3_out = self.conv3(relu2_out) relu4_out = self.relu4(conv3_out) return relu4_out
You are welcome (and encouraged) to use the built-in batch normalization and dropout layer.
Guessing that the pixel is not cancerous every single time will give you an accuracy of ~ 85%. Your trained network should be able to do better than that (but you will not be graded on accuracy). This is the result I got after 1 hour or training.