This is an old revision of the document!
Table of Contents
Objective:
To understand expectation maximization, and to explore how to learn the parameters of a Gaussian mixture model.
Deliverable:
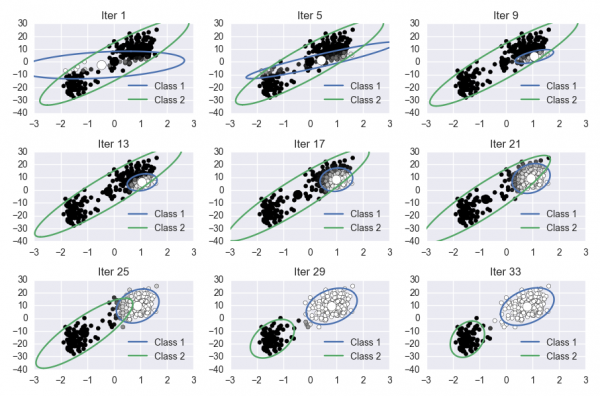
For this lab, you will implement the Expectation Maximization algorithm on the Old Faithful dataset. This involves learning the parameters of a Gaussian mixture model. Your notebook should produce a visualization of the progress of the algorithm. The final figure should look something like this:
Grading:
Your notebook will be graded on the following elements:
- 10% Data is correctly mean-centered
- 20% Correctly updates responsibilities
- 20% Correctly updates means
- 20% Correctly updates covariances
- 20% Correctly updates mixing weights
- 10% Final plot is tidy and legible
Description:
For this lab, you will use the Old Faithful dataset, which you can download here:
To help our TA better grade your notebook, you should use the following initial parameters:
# the Gaussian means (as column vectors -- ie, the mean for Gaussian 0 is mus[:,0] mus = np.asarray( [[-1.17288986, -0.11642103], [-0.16526981, 0.70142713]]) # the Gaussian covariance matrices covs = list() covs.append( np.asarray([[ 0.74072815, 0.09252716], [ 0.09252716, 0.5966275 ]]) ) covs.append( np.asarray([[ 0.39312776, -0.46488887], [-0.46488887, 1.64990767]]) ) # The Gaussian mixing weights mws = [ 0.68618439, 0.31381561 ]
Hints:
In order to visualize a covariance matrix, you should plot an ellipse representing the 95% confidence bounds of the corresponding Gaussian. Here is some code that accepts as input a covariance matrix, and returns a set of points that define the correct ellipse; these points can be passed directly to the plt.plot() command as the x and y parameters.
import numpy as np def cov_to_pts( cov ): circ = np.linspace( 0, 2*np.pi, 100 ) sf = np.asarray( [ np.cos( circ ), np.sin( circ ) ] ) [u,s,v] = np.linalg.svd( cov ) pmat = u*2.447*np.sqrt(s) # 95% confidence return np.dot( pmat, sf )
Here are some additional python functions that may be helpful to you:
# compute the likelihood of a multivariate Gaussian scipy.stats.multivariate_normal.pdf # scatters a set of points; check out the "c" keyword argument to change color, and the "s" arg to change the size plt.scatter plt.xlim # sets the range of values for the x axis plt.ylim # sets the range of values for the y axis # to check the shape of a vector, use the .shape member foo = np.random.randn( 100, 200 ) foo.shape # an array with values (100,200) # to transpose a vector, you can use the .T operator foo = np.atleast_2d( [42, 43] ) # this is a row vector foo.T # this is a column vector import numpy as np np.atleast_2d np.sum